One of the interests of Siegfried Musser group from Texas AM University is to study molecular transport via Nuclear Pore Complexes (NPCs) across the nuclear membrane inside the cell. As a key tool in their research, scientists are using Single Molecule Localization Microscopy (SMLM) boosted by Adaptive Optics (AO), which provides increased resolution both in time and space (localization precision).
Recently, a second paper was published on this topic, providing a systematic analysis of the influence of some key imaging parameters – including AO – on the quality of SMLM images.
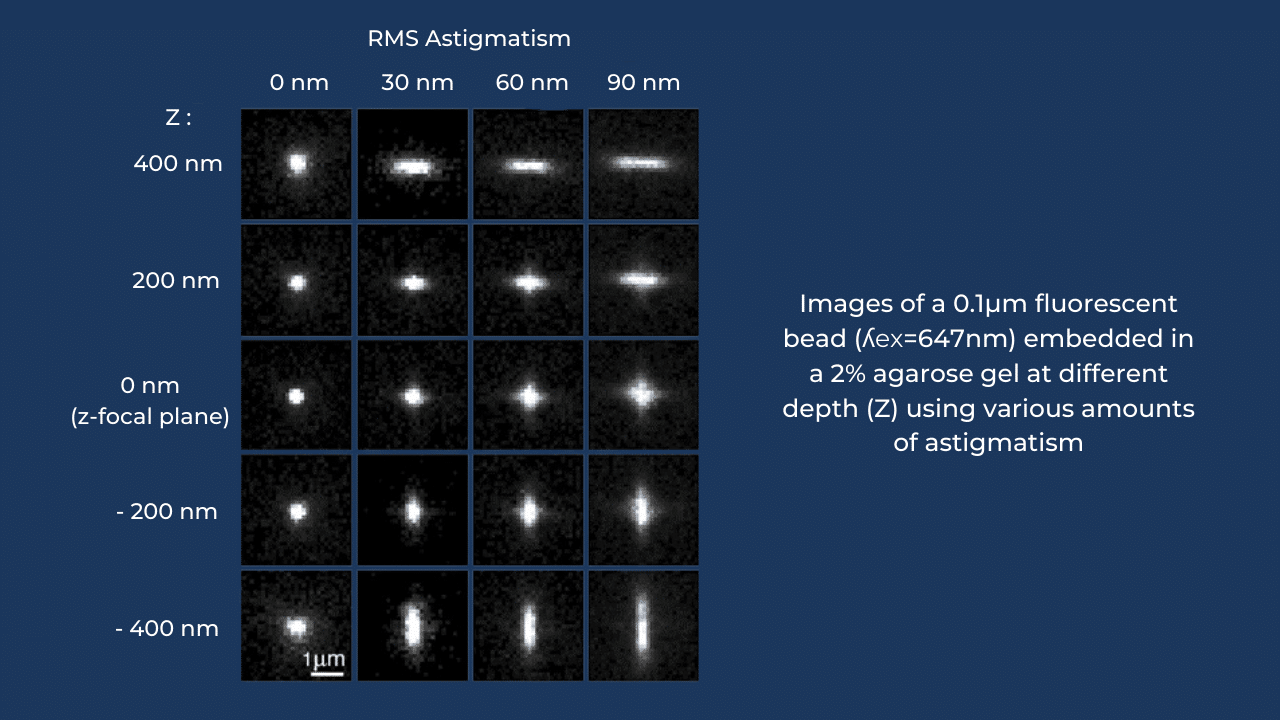
Astigmatism is probably the simplest and most efficient PSF shaping method, allowing to systematically reach localization precisions around 5-10nm @4000 arriving photons (a typical amount in real-life situation). Astigmatic PSF can be obtained by simply adding a cylindrical lens to the emission pathway of the microscope. But the amplitude of astigmatism in this case is fixed. Another way to induce astigmatism is by using a deformable mirror, which enables to correct aberrations and to introduce pure astigmatism with an amplitude that can be instantaneously varied over a large range.
In this paper, scientists carefully evaluated the following factors influencing both lateral and axial localization precision: the number of incident photons, the amplitude of induced astigmatism and even the type of acquisition camera (EMCCD and sCMOS). As for AO, it was shown that by changing the amount of induced astigmatism it is possible to fine tune the ratio of lateral and axial localization precision. A homogeneous localization precision is obtained when using 90nm RMS of astigmatism on the deformable mirror: it allows reaching localization precision of about 10nm in all three dimensions at 3000 incident photons.
In this study scientists used AOKit Bio product bundle, which is composed of Mirao 52E deformable mirror, HASO4 First wavefront sensor for mirror calibration, and an adaptive optics software including a 3N image-based sensorless aberration correction process. The implementation design of AOKit Bio is described in this paper to a great detail. For easier, faster integration of the same AO approach, our add-on device MicAO 3DSR can be used, enabling easy implementation on any inverted frame microscope.
