In their quest to better understand the disassembly of NPCs during mitosis, the team of Prof. Ulrike Kutay at Zurich ETH decided to dig deeper into Y-complex behaviour.
Why observe Y-complexes?
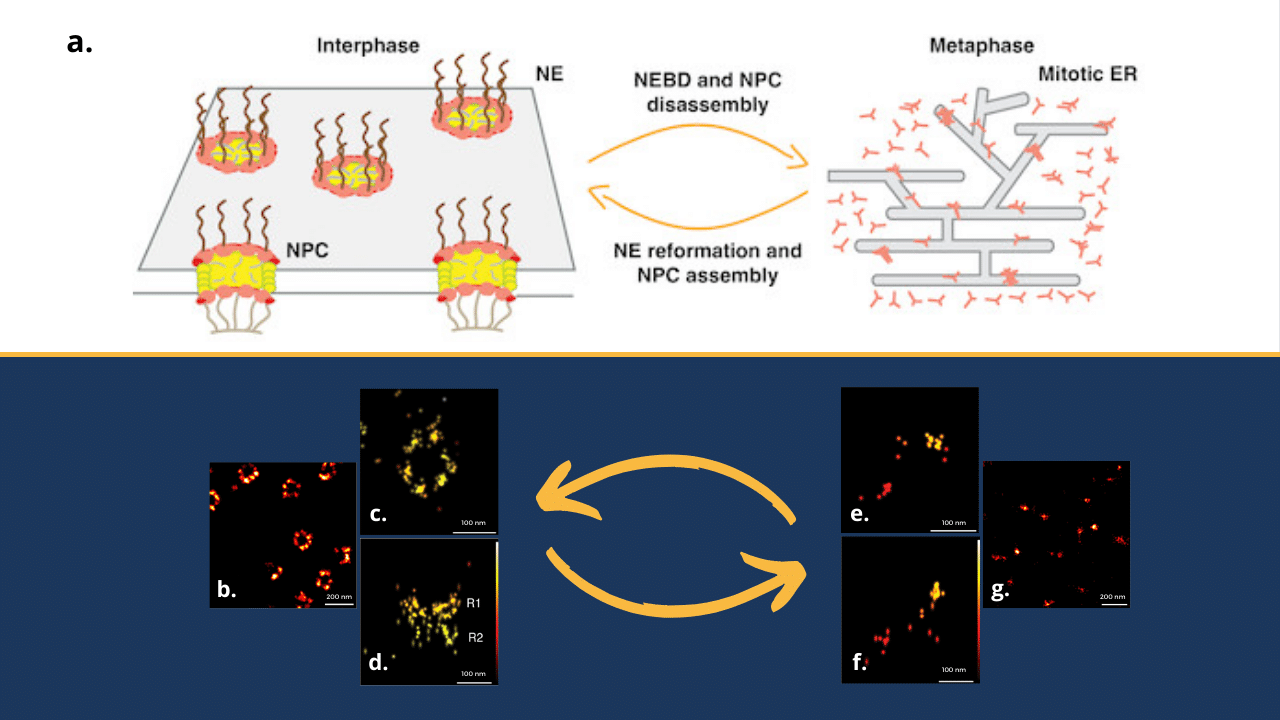
During mitotic entry, the dismantling of NPCs seems to be a two-tier process and if disassembly of most components occurs rapidly, Y-complex Nups and cytoplasmic rings display slower dissociation kinetics, indicating a mechanistic difference in their splitting.
The challenge here? Observing such small cellular elements requires sufficient optical resolution, going below 20nm laterally and 50nm axially. This is where single molecule localization microscopy (SMLM) in a combination with adaptive optics (AO) comes into play.
Single Molecule Localization Microscopy for a better lateral resolution
First, let’s see what SMLM can bring. This group of imaging techniques enable scientists to breach the conventional diffraction limit, allowing them to visualize structures at the nanoscale. The diverse methods that are regrouped under the umbrella term SMLM are using the properties of some fluorophores to switch from dark to emissive state and vice-versa. In this case, Prof. Kutay team used dSTORM, a method allowing to capture details on the order of tens of nanometres – a sufficient resolution to observe Y-complexes in both interphasic (image b) and mitotic (image g) states in lateral dimension. But what about 3D?
Playing with astigmatism to get an axial dimension
While SMLM offers remarkable lateral resolution, understanding the complete three-dimensional architecture of Y-complexes requires tackling the axial dimension. One of ways to address this question is the shaping of point spread function (PSF) with astigmatism using adaptive optics. With this method, researchers were able to reach localization precision around 5-10nm and obtained 3D images of Y-complexes in their ring shape (during interphase, images c and d) and in their ER-bound shape (during mitosis, images e and f).
How can research benefit from adaptive optics?
In conclusion, boosting your SMLM system with adaptive optics will help you to see new details in cellular structures, complexes and molecules, just as Prof. Kutay group did (read their full article here).
But how to implement adaptive optics? Well, in their paper, researchers were using MicAO 3DSR device. This adaptive optics module, which is simply inserted between the microscope and imaging camera, can indeed enable you to perform 3D SMLM using astigmatism as mentioned above. If adaptive optics are a real game changer for 3D super-resolution microscopy, it is also a very powerful tool for 2D. Indeed, MicAO 3DSR can correct sample-induced aberrations and help increasing photon collection, resulting in increase of localization precision that, in the end, will allow you to obtain the highest quality super resolved 3D structures.
You have questions regarding adaptive optics in microscopy? Do not hesitate to contact us!
